Model Uncertainty Estimation in Interatomic Potentials
Large scale molecular dynamics (MD) simulations rely on interatomic potentials that are pre-constructed using ab initio data as well as both empirical and physical considerations. Machine-learned interatomic potentials (MLIAPs) are fitted to data available from expensive quantum chemistry computations. Spectral neighbor analysis potential (SNAP) is a special class of linearly parameterized potentials with bispectrum feature set, developed and implemented in Sandia’s FitSNAP software.
Uncertainty quantification (UQ) for MLIAPs is critical for both training data selection and model selection. Besides, MLIAPs equipped with UQ enable the propagation of uncertainty through MD simulations, thereby providing uncertainty estimates on MD simulation outputs and allowing for meaningful comparisons with experimentally observed macroscale quantities of interest.
We implemented several UQ approaches, including embedded model error [1], as UQ-solvers within FitSNAP [2], providing model uncertainty estimation of the learned interatomic potentials. Besides, we have applied the learned uncertainty in an active learning outer-loop to enable efficient selection of training configurations [3].
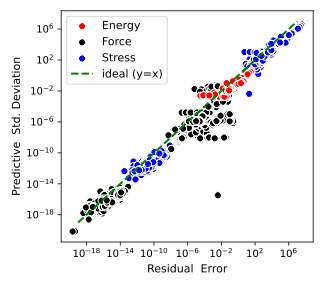
FigureBayesian predictive uncertainty of SNAP potential is well-correlated with the residual error, enabling efficient active learning.
AcknowledgementsThe work is supported by DOE Office of Science ASCR and FES programs.
References[1] C. Lacey, K. Sargsyan, H. N. Najm, In prep., 2024.
[2] A. Rohskopf et al., “FitSNAP: Atomistic machine learning with LAMMPS”, Journal of Open Source Software, 8(84), 5118, 2023.
[3] L. Williams, K. Sargsyan, A. Rohskopf, H. N. Najm, “Active learning for SNAP interatomic potentials via Bayesian predictive uncertainty”, Computational Materials Science, 242, p113074, 2024.